Python in itself natively executes single-threaded. There are libraries that allow the possibility of executing code multi-threaded but it involves complexities. The other downside is the code doesn't scale well enough to the number of execution threads (or cores) the code runs on. Running single-threaded code is stable and proven, but it just takes a while to execute.
I have been on the receiving end of the single-threaded execution. It takes a while to execute, and during the development stage the workaround is to slice a sample of the dataset so that execution does not have to take a long time. More often than not, this is acceptable. Recently, I stumbled on a setup that takes code and executes Python multi-threaded. What is cool about it? It scales to the number of cores thrown at it, and it scales to other nodes as well (think distributed computing).
This is particularly applicable to the field of data science and analytics, where the datasets grow into the hundreds of millions and even billions of rows of data. And since Python is the code of choice in this field, PySpark shines. I need not explain the details of PySpark as a lot of resources already do that. Let me describe the set-up so that code executes in as many cores as you can afford.
The derived procedure is based on an Ubuntu 16 LTS installed on a VirtualBox hypervisor, but is very repeatable whether the setup is in Amazon Web Services (AWS), Google Cloud Platform (GCP) or your own private cloud infrastructure, such as VMware ESXi.
Please note that the procedure will enclose the commands to execute in [square brackets]. Start by updating the apt repository with the latest packages [sudo apt-get update]. Then install scala [sudo apt-get -y install scala]. In my experience this installs the package "default-jre" but in case it doesn't, install default-jre as well [sudo apt-get -y install default-jre].
Download miniconda from the continuum repository. On the terminal, execute this command [wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh]. This link points to the 64-bit version of python3. Avoid python2 as much as possible, since development for it is approaching its end; 64-bit is almost always the default. Should you want to install the heavier anaconda3 in place of miniconda3, you may opt to do so.
Install miniconda3 [bash Miniconda3-latest-Linux-x86_64.sh] on your home directory. This avoids package conflicts with the pre-packaged python of the operating system. At the end of the install, the script will ask to modify the PATH environment to the installation directory. Accept the default option, which to modify the PATH. This step is optional, but if you want to you may add the conda-forge channel [conda config --add channels conda-forge] in addition to the default base channel.
At this point, the path where miniconda was installed needs to precede the path where the default python3 resides [source $HOME/.bashrc]. This of course assumes that you chose to accept .bashrc modification as suggested by the installer. Next, use conda to install py4j and pyspark [conda install --yes py4j pyspark]. The install will take a while so go grab some coffee first.
While the install is taking place, download the latest version of spark. As of this writing, the latest version is 2.2.1 [wget http://mirror.rise.ph/apache/spark/spark-2.2.1/spark-2.2.1-bin-hadoop2.7.tgz]. Select a download mirror that is closer to your location. Once downloaded unpack the tarball on your home directory [tar zxf spark-2.2.1-bin-hadoop2.7.tgz]. A directory named "spark-2.2.1-bin-hadoop2.7" will be created in your home directory. It contains the binaries for spark. (This next step is optional, as this is my personal preference.) Create a symbolic link to the directory "spark-2.2.1-bin-hadoop2.7" [ln -s spark-2.2.1-bin-hadoop2.7 spark].
The extra step above will make things easier to upgrade spark (since spark is actively being developed). Simply re-point spark to the newly unpacked version without having to modify the environment variables. If there are issues with the new version, simply link "spark" back to the old version. Think of it like a switch with the clever use of a symbolic link.
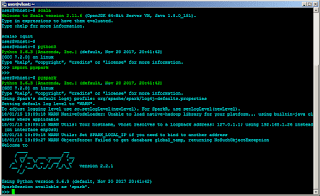
At this point, all the necessary software are installed. It is imperative that checks are done to ensure that the software are working as expected. For scala, simply run [scala] without any options. If you see the welcome message, it is working. For pyspark, either import the pyspark library in python [import pyspark] or execute [pyspark] on the terminal. You should see a similar screen as below.
Modify the environment variables to include SPARK_HOME [export SPARK_HOME=$HOME/spark]. Make changes permanent by putting that in ".bashrc" or ".profile". Likewise, add $HOME/spark/bin to PATH.
RELATED: Data Science -- Where to Start?
This setup becomes even more robust by integrating pyspark with the jupyter notebook development environment. This is a personal preference and I will cover that in a future post.
Subscribe for Latest Update
Popular Posts
-
I have been a Dell Latitude D630 user for over 3 years now. I have used it in my previous job; I use one personally; and I have been assigne...
-
News from around the web recently revolved around security breaches and hacked websites. And when news broke out, many others found out abou...
-
Windows 7 SP1 has just gone live over a few days ago. If you haven't gone and downloaded the Service Pack yet, you may check it out here...
-
I could say I'm too familiar with computers to confidently install just about any modern Windows or Linux operating system from USB or b...
-
I was setting up CentOS7 in order to orient myself with SELinux and be comfortable with using it. If you have been an avid follower of my ar...
Post Labels
100gb
(1)
acceleration
(1)
acrobat
(1)
adblock
(1)
advanced
(1)
ahci
(1)
airdrop
(2)
aix
(14)
angry birds
(1)
article
(21)
aster
(1)
audiodg.exe
(1)
automatic
(2)
autorun.inf
(1)
bartpe
(1)
battery
(2)
bigboss
(1)
binance
(1)
biometrics
(1)
bitcoin
(3)
blackberry
(1)
book
(1)
boot-repair
(2)
calendar
(1)
ccleaner
(3)
chrome
(5)
cloud
(1)
cluster
(1)
compatibility
(3)
CPAN
(1)
crypto
(3)
cydia
(1)
data
(3)
ddos
(1)
disable
(1)
discount
(1)
DLNA
(1)
dmidecode
(1)
dns
(7)
dracut
(1)
driver
(1)
error
(10)
esxi5
(2)
excel
(1)
facebook
(1)
faq
(36)
faucet
(1)
firefox
(17)
firewall
(2)
flash
(5)
free
(3)
fun
(1)
gadgets
(4)
games
(1)
garmin
(5)
gmail
(3)
google
(4)
google+
(2)
gps
(5)
grub
(2)
guide
(1)
hardware
(6)
how
(1)
how-to
(45)
huawei
(1)
icloud
(1)
info
(4)
iphone
(7)
IPMP
(2)
IPV6
(1)
iscsi
(1)
jailbreak
(1)
java
(3)
kodi
(1)
linux
(28)
locate
(1)
lshw
(1)
luci
(1)
mafia wars
(1)
malware
(1)
mapsource
(1)
memory
(2)
mikrotik
(5)
missing
(1)
mods
(10)
mouse
(1)
multipath
(1)
multitasking
(1)
NAT
(1)
netapp
(1)
nouveau
(1)
nvidia
(1)
osmc
(1)
outlook
(2)
p2v
(2)
patch
(1)
performance
(19)
perl
(1)
philippines
(1)
php
(1)
pimp-my-rig
(9)
pldthomedsl
(1)
plugin
(1)
popcorn hour
(10)
power shell
(1)
process
(1)
proxy
(2)
pyspark
(1)
python
(13)
qos
(1)
raspberry pi
(7)
readyboost
(2)
reboot
(2)
recall
(1)
recovery mode
(1)
registry
(2)
rename
(1)
repository
(1)
rescue mode
(1)
review
(15)
right-click
(1)
RSS
(2)
s3cmd
(1)
salary
(1)
sanity check
(1)
security
(15)
sendmail
(1)
sickgear
(3)
software
(10)
solaris
(17)
squid
(3)
SSD
(3)
SSH
(9)
swap
(1)
tip
(4)
tips
(42)
top list
(3)
torrent
(5)
transmission
(1)
treewalk
(2)
tunnel
(1)
tweak
(4)
tweaks
(41)
ubuntu
(4)
udemy
(6)
unknown device
(1)
updates
(12)
upgrade
(1)
usb
(12)
utf8
(1)
utility
(2)
V2V
(1)
virtual machine
(4)
VirtualBox
(1)
vmware
(14)
vsphere
(1)
wannacry
(1)
wifi
(4)
windows
(54)
winpe
(2)
xymon
(1)
yum
(1)
zombie
(1)
RANDOM POSTS
-
the posts here are my own and not shared nor endorsed by the companies i am affiliated with.
i am a technologist who likes to automate and make things better and efficient. i can be reached via great [dot] dilla [at] gmail [dot] com.
if this post has helped, will you buy me coffee?